GENESIS Tutorial 9.2 (2022)
Restraints in the NPT ensemble
In this tutorial, we will describe how to deal with positional restraints in the NPT ensemble. Our target biological system here is a GPCR system, and we investigate the effect of virial calculation from positional restraint in performing NPT simulations.
1. Preparation
All the files required for this tutorial are hosted in the GENESIS tutorials repository on GitHub. If you haven’t downloaded the files yet, open your terminal and run the following command (see more in Tutorial 1.1):
$ cd ~/GENESIS_Tutorials-2022
# if not yet
$ git clone https://github.com/genesis-release-r-ccs/genesis_tutorial_materials
If you already have the tutorial materials, let’s go to our working directory:
$ cd genesis_tutorial_materials/tutorial-11.1
$ ls
1.min 2.equil_pressure_virial step5_input.pdb toppar
2.equil restraints step5_input.psf
System and force fields information are described in toppar, restraints,
step5_input.pdb, and step5_input.psf.
2. Minimization
Let’s consider the first step (step 1) in our tutorial: minimization. The
input file for minimization is in the 1.min directory. Please run the
minimization with the following command:
$ cd 1.min
$ mpirun -np 8 {PATH}/spdyn inp > out
Here, we assign positional restraints in three selection groups: (1) heavy backbone atoms, (2) heavy sidechain atoms, and (3) phosphate atoms in POPC lipids. They are written as:
# Selection group
[SELECTION]
group1 = (sid:PROA) and backbone
group2 = (sid:PROA) and not backbone and not hydrogen
group3 = sid:MEMB and ((rnam:POPC and (an:P)))
# Positional restraint function
[RESTRAINTS]
nfunctions = 3
# Restraint function of the first group
function1 = POSI
constant1 = 10.0
select_index1 = 1
# Restraint function of the second group
function2 = POSI
constant2 = 5.0
select_index2 = 2
# Restraint function of the third group
function3 = POSI
direction3 = Z
constant3 = 2.5
select_index3 = 3
For more information on how to setup the rest of the options, please see Tutorial 3.2.
3. Equilibration
We perform equilibration with two different conditions: with and without pressure from positional
restraint force. In 2.equil, the virial contribution from the positional restraint force is not
considered in the pressure evaluation of the NPT simulations (It is the default in GENESIS 1.X and 2.0).
On the other hand, in 2.equil_pressure_virial, virial from positional restraint force is included.
3.1. Equilibration without positional restraint contribution to pressure
In the equilibration run, we are considering the following steps:
- step 2: NVT simulation with 1 fs time step (125000 steps)
- step 3: NVT simulation with 1 fs time step (125000 steps)
- step 4: NPT simulation with 1 fs time step (125000 steps)
- step 5: NPT simulation with 2 fs time step (250000 steps)
- step 6: NPT simulation with 2 fs time step (250000 steps)
- step 7: NPT simulation with 1 fs time step (25000 steps)
In 2.equil, we can see input files as following:
$ cd 2.equil
$ ls
box.py inp10 inp12 inp2 inp4 inp6 inp8
inp1 inp11 inp13 inp3 inp5 inp7 inp9
inp1 to inp6 are input files for step2 to step 7, respectively. They all have positional restraint functions on the same three groups previously selected in the minimization. The restraint forces are weaken as we move on the next step.
step 2: NVT simulation with 1 fs time step
$ ls inp1
# restraint function in input file inp1
...
[RESTRAINTS]
nfunctions = 3
function1 = POSI
constant1 = 10.0
select_index1 = 1
function2 = POSI
constant2 = 5.0
select_index2 = 2
function3 = POSI
direction3 = Z
constant3 = 2.5
select_index3 = 3
step 3: NVT simulation with 2 fs time step
$ ls inp2
# restraint function in input file inp2
...
[RESTRAINTS]
nfunctions = 3
function1 = POSI
constant1 = 5.0
select_index1 = 1
function2 = POSI
constant2 = 2.5
select_index2 = 2
function3 = POSI
direction3 = Z
constant3 = 2.5
select_index3 = 3
step 4: NPT simulation with 1 fs time step
$ ls inp3
# restraint function in input file inp3
...
[RESTRAINTS]
nfunctions = 3
function1 = POSI
constant1 = 2.5
select_index1 = 1
function2 = POSI
constant2 = 1.0
select_index2 = 2
function3 = POSI
direction3 = Z
constant3 = 1.0
select_index3 = 3
step 5: NPT simulation with 2 fs time step
$ ls inp4
# restraint function in input file inp4
...
[RESTRAINTS]
nfunctions = 3
function1 = POSI
constant1 = 1.0
select_index1 = 1
function2 = POSI
constant2 = 0.5
select_index2 = 2
function3 = POSI
direction3 = Z
constant3 = 0.5
select_index3 = 3
step 6: NPT simulation with 2 fs time step
$ ls inp5
# restraint function in input file inp5
...
[RESTRAINTS]
nfunctions = 3
function1 = POSI
constant1 = 0.5
select_index1 = 1
function2 = POSI
constant2 = 0.1
select_index2 = 2
function3 = POSI
direction3 = Z
constant3 = 0.1
select_index3 = 3
step 7: NPT simulation with 1 fs time step
$ ls inp6
# restraint function in input file inp6
...
[RESTRAINTS]
nfunctions = 3
function1 = POSI
constant1 = 0.1
select_index1 = 1
function2 = POSI
constant2 = 0.0
select_index2 = 2
function3 = POSI
direction3 = Z
constant3 = 0.0
select_index3 = 3
You can perform these by executing genesis with each input file:
$ cd 2.equil
$ mpirun -np 8 {PATH}/spdyn inp1 > out1 #step 2
$ mpirun -np 8 {PATH}/spdyn inp2 > out2 #step 3
$ mpirun -np 8 {PATH}/spdyn inp3 > out3 #step 4
$ mpirun -np 8 {PATH}/spdyn inp4 > out4 #step 5
$ mpirun -np 8 {PATH}/spdyn inp5 > out5 #step 6
$ mpirun -np 8 {PATH}/spdyn inp6 > out6 #step 7
3.2. Equilibration with positional restraint contribution to pressure
We do the same equilibration procedure with positional restraint contribution to pressure in 2.equil_pressure_virial:
$ cd 2.equil_pressure_virial
$ mpirun -np 8 {PATH}/spdyn inp1 > out1 #step 2
$ mpirun -np 8 {PATH}/spdyn inp2 > out2 #step 3
$ mpirun -np 8 {PATH}/spdyn inp3 > out3 #step 4
$ mpirun -np 8 {PATH}/spdyn inp4 > out4 #step 5
$ mpirun -np 8 {PATH}/spdyn inp5 > out5 #step 6
$ mpirun -np 8 {PATH}/spdyn inp6 > out6 #step 7
Input files from inp1 to inp6 are the same as those in 2.equil except for one thing: to allow pressure evaluation from positional restraint force, we write
pressure_position = YES
in the [RESTRAINTS] section.
4. Production and Analysis
After finishing the equilibration, we perform the produciton run without positional restraints (step 8). It can be done by running genesis with inp7 and inp8. In the case of 2.equil, the produciton run is done by the following commands:
$ cd 2.equil
$ mpirun -np 8 {PATH}/spdyn inp7 > out7
$ mpirun -np 8 {PATH}/spdyn inp8 > out8
Similary, we can do the same thing in 2.equil_pressure_virial:
$ cd 2.equil_pressure_virial
$ mpirun -np 8 {PATH}/spdyn inp7 > out7
$ mpirun -np 8 {PATH}/spdyn inp8 > out8
Finally, we investigate system size changes (x and z dimensions) during the equilibration and production steps. In each directory, 2.equil and 2.equil_pressure_virial, please execute the python script:
$ python box.py > box.dat
In the file, box.dat, the first, second, and third columns represent time (ps), system size in x dimension, and system size in the z dimension, respectively. In the case of 2.equil, the system size as a function of the MD simulation time is shown as
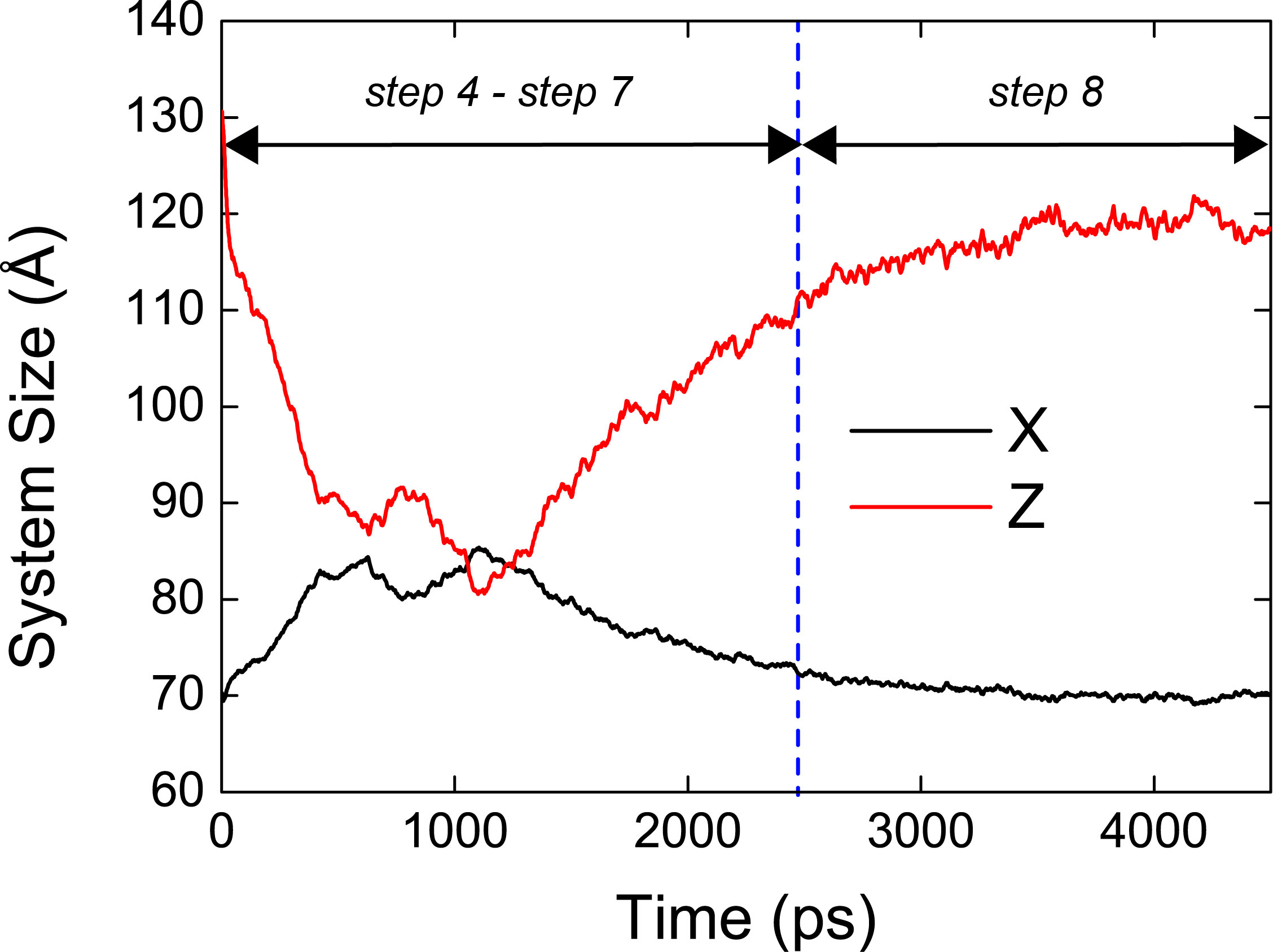
We can easily understand that there is a large change of the system size during the equilibration and production. This means that the system is not well equilibrated when we neglect pressure contribution from positional restraint forces. Unlike 2.equil, the system size does not change during the equilibration and production steps in 2.equil_pressure_virial.
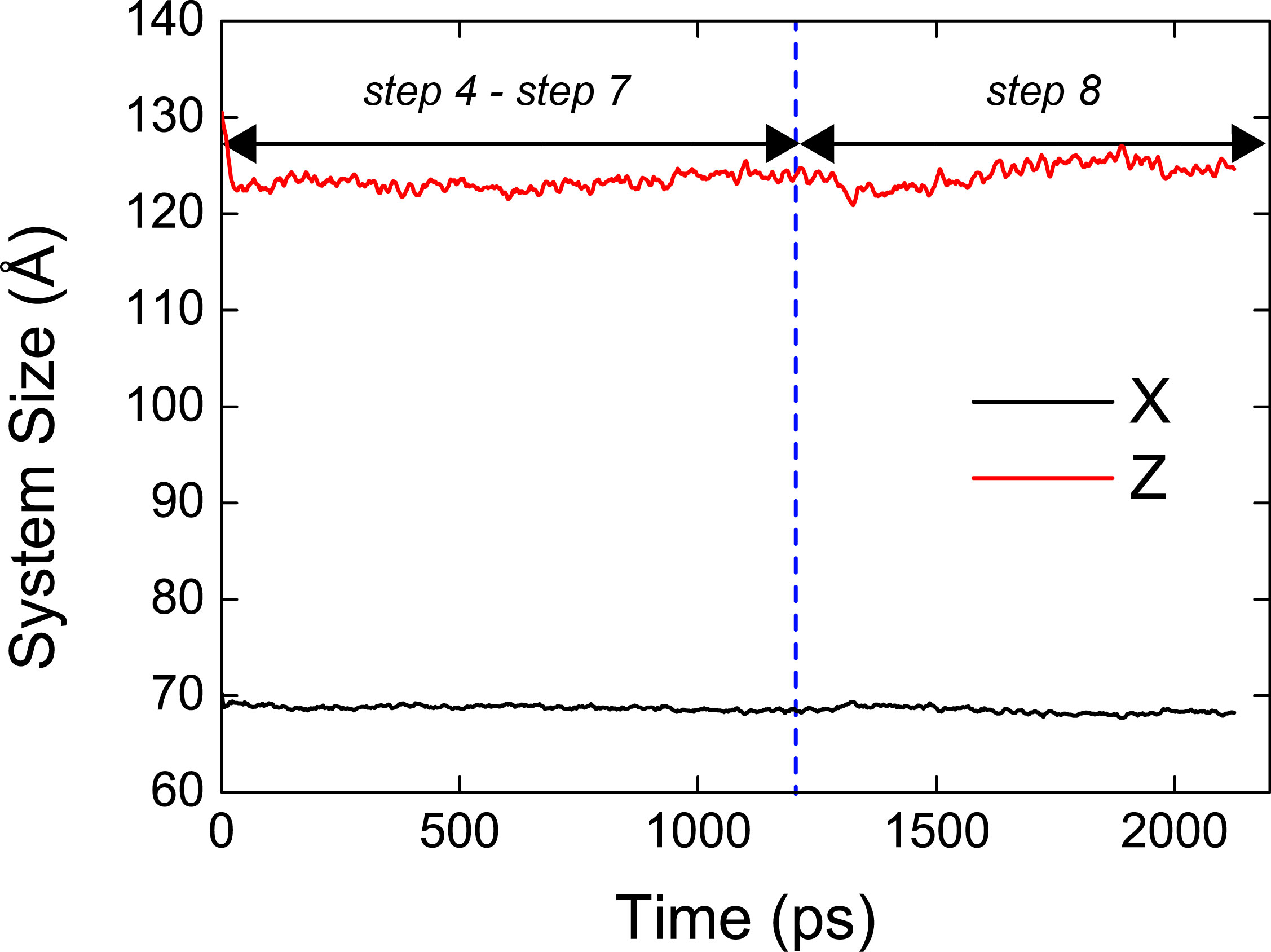
Therefore, we should be careful when performing NPT simulations with
positional restraints. To obtain a reliable structure, we recommend
including the virial contribution from the positional restraints by
writing pressure_position = YES. Similarly, we recommend writing
pressure_rmsd = YES when performing NPT simulations with RMSD
restraints.
Written by Jaewoon Jung@RIKEN R-CCS. March, 2022
Updated by Jaewoon Jung@RIKEN R-CCS. May, 16, 2022