GENESIS Tutorial 11.3 (2022)
Coarse-grained MD simulation of TBP-DNA Sequence-Specific Recognition
Notice: This tutorial is for GENESIS v1.7.0 and later!
In this tutorial we will simulate the specific recognition of a special DNA sequences, the “TATA-box”, by the TATA-box binding protein (TBP). TBP is a general transcription factor that participates in the initialization of transcription. It uses a beta-sheet surface to bind to the minor groove of DNA and induces sharp bending of DNA. Here we will use the AICG2+ model1 for TBP and the 3SPN.2C model2 for DNA. For intermolecular protein-DNA interactions, we consider excluded-volume, electrostatics, and sequence-specific interactions. Particularly, the last one is modeled by the PWMcos method.3
0. Preparations
0.1 Install necessary softwares
Same as in tutorial 11.1 and 11.2, we will use the GENESIS-CG-tool 4 to generate CG coordinate and topology files.
0.2 Download the files for this tutorial
All the files required for this tutorial are hosted in the GENESIS tutorials repository on GitHub.
If you haven’t downloaded the files yet, open your terminal and run the following command (see more in Tutorial 1.1):
$ cd ~/GENESIS_Tutorials-2022
# if not yet
$ git clone https://github.com/genesis-release-r-ccs/genesis_tutorial_materials
If you already have the tutorial materials, let’s go to our working directory:
$ cd genesis_tutorial_materials/tutorial-11.3
This tutorial consists of two parts: 1) system setup; and 2) MD simulations of TBP binding on DNA:
$ ls
01_setup 02_simulation
1. Setup
1.1 Prepare the DNA structure and topology
Let’s go to our working directory:
$ cd 01_setup
$ ls
dsDNA.fasta
Here we want to simulate the binding of TBP on a 50bp dsDNA. The DNA
sequence file dsDNA.fasta looks like this:
> dsDNA for TBP binding
AGCAATTAGCCAGGGAATGTATAAAAGGCGTCAGGGAGACTCACTGGGCT
Here we will prepare the DNA topology and coordinate files in the same way as in tutorial 11.2:
$ /home/user/genesis_cg_tool/tools/modeling/DNA_general/build_dna.jl -s dsDNA.fasta -C -o dsDNA
Please refer to tutorial 11.2 for more details of generating DNA CG files.
1.2 Prepare the protein topology files
Now let’s download the PDB file from RCSB:
$ wget https://files.rcsb.org/download/1CDW.pdb
We first make a copy of the PDB file and do some modification so that it contains only the protein.
$ grep -E "^ATOM.{17}A" 1CDW.pdb > tbp.pdb
$ echo "END" >> tbp.pdb
Now let’s generate the CG topology and coordinate fils for TBP:
$ /home/user/genesis_cg_tool/src/aa_2_cg.jl tbp.pdb
We will get several new files: tbp_cg.gro, tbp_cg.top, and tbp_cg.itp. The
first one is the coordinate file for TBP, and the later two are the topology
files.
So far we have finished the preparation for the DNA and the protein, respectively. The commands have been also introduced in tutorial 11.1 and 11.2. However, these files are not enough to study the specific recognition between TBP and DNA. In the next section we will show how to prepare the sequence-specific interaction parameters.
1.3 Introduce sequence specificity
In the PWMcos model, sequence specificity is described by the position weight matrix (PWM). However, very often another form, the position frequency matrix (PFM), is provided by experiments. The PFMs are deposited on databases like JASPAR and UniPROBE. Now let’s download the PFM from the TBP entry on JASPAR.
$ wget http://jaspar.genereg.net/api/v1/matrix/MA0108.1.jaspar
The PFM file for TBP looks like this:
> MA0108.1 TBP
A [ 61 16 352 3 354 268 360 222 155 56 83 82 82 68 77 ]
C [ 145 46 0 10 0 0 3 2 44 135 147 127 118 107 101 ]
G [ 152 18 2 2 5 0 10 44 157 150 128 128 128 139 140 ]
T [ 31 309 35 374 30 121 6 121 33 48 31 52 61 75 71 ]
Each element in the matrix represents the frequency of a base type (“A”, “C”, “G”, or “T”) appearing at a position. The matrix also shows the “consensus sequence”, which is the most probable target sequence. Based on the PFM above, the consensus sequence for TBP is: “GTATAAAAGGCGGGG”.
The PWMcos model incorporates the PFM and the protein-DNA binding structural information based on the PDB structure. For the model to work, we have to match up the consensus sequence with the DNA sequence in the TBP-DNA complex PDB structure (1CDW). Note that for some proteins the available PDB DNA sequence can be different from the PFM consensus sequence. Therefore, we have to add the sequence index information into the PFM file, and then use our GENESIS-CG-tool to extract CG parameters.
Let’s first use the GENESIS-CG-tool to get the sequence information from “1CDW.pdb”:
$ /home/user/genesis_cg_tool/src/aa_2_cg.jl 1CDW.pdb --show-sequence
This command generates a new file, 1CDW_cg.fasta, which lists the
sequence of dsDNA and TBP:
> Chain A : DNA
CTGCTATAAAAGGCTG
> Chain B : DNA
CAGCCTTTTATAGCAG
> Chain C : protein
SGIVPQLQNIVSTVNLGCKLDLKTIALRARNAEYNPKRFAAVIMRIREPRTTALIFSSGKMVCTGAKSEENSRLAARKYARVVQKLGFPAKFLDFKIQNMVGSCDVKFPIRLEGLVLTHQQFSSYEPELFPGLIYRMIKPRIVLLIFVSGKVVLTGAKVRAEIYEAFENIYPILKGFRK
For clarity, here we align the PFM consensus sequence and the sequence
of chain A in the PDB, as shown below:

As can be seen in the figure, the “TATAAAAG” motif matches a segment of “Chain A”. We then add the indices of each base to a new “PFM” file:
$ cp MA0108.1.jaspar tbp.pfm
$ vim tbp.pfm
...
This is the content of the new file tbp.pfm:
$ cat tbp.pfm
A 61 16 352 3 354 268 360 222 155 56 83 82 82
C 145 46 1 10 1 1 3 2 44 135 147 127 118
G 152 18 2 2 5 1 10 44 157 150 128 128 128
T 31 309 35 374 30 121 6 121 33 48 31 52 61
CHAIN_A 4 5 6 7 8 9 10 11 12 13 14 15 16
CHAIN_B 29 28 27 26 25 24 23 22 21 20 19 18 17
Compared with the original PFM file, we added two more lines, “CHAIN_A” and “CHAIN_B”, listing the indices of each base (in CHAIN_A) and its complementary base (in CHAIN_B) in the PDB file, respectively. Let’s take column 3 (16, 46, 18, 309, 5, and 28) as an example, the first four elements, 16, 46, 18, and 309 show the frequency of each base type appearing at this position, from which we can tell that the most probable base-pair at this position is “T-A”. Correspondingly, the last two elements in this column, 5 and 28, describe the residue indices of the “T” and “A” in PDB, respectively. Note that some elements in the PFM are changed from “0” to “1” (for instance, row 2 column 3 of base type “C”). This is because in the transition from PFM to PWM, we have to take the logarithm of each element of the PFM. Therefore, we replace all the “0”s in the PFM with some small “pseudo counts”.5
It’s time to run the GENESIS-CG-tool again:
$ /home/user/genesis_cg_tool/src/aa_2_cg.jl 1CDW.pdb --pwmcos -p tbp.pfm --patch tbp_cg.itp --pwmcos-scale 3.9 --pwmcos-shift -0.4
Here the --pwmcos option tell the program to generate parameters for the
PWMcos model, -p specifies the PFM file, and the --patch option is used to
tell GENESIS-CG-tool to append information to the .itp file, instead of writing
a new file. The options --pwmcos-scale and --pwmcos-shift are used to
specify energy scaling and shifting factor parameters (see reference 3).
Let’s take a look at the PWMcos information added to the end of tbp_cg.itp:
$ tail -n 34 tbp_cg.itp
[ pwmcos ]
; i f r0 theta1 theta2 theta3 ene_A ene_C ene_G ene_T gamma eps'
9 1 0.84956 57.727 50.211 73.212 -0.549774 0.649174 0.649174 -0.748573 3.900 -0.400
9 1 0.71936 43.555 90.626 100.377 -0.136243 0.222109 0.543997 -0.629863 3.900 -0.400
11 1 0.73831 74.045 74.387 97.686 -0.748573 0.649174 0.649174 -0.549774 3.900 -0.400
11 1 0.77045 77.501 119.188 66.900 -0.779085 0.417788 0.116795 0.244501 3.900 -0.400
11 1 0.84433 73.607 95.508 117.177 -0.549774 0.649174 0.649174 -0.748573 3.900 -0.400
39 1 0.89446 88.977 80.059 46.823 -0.698333 0.560902 -0.711154 0.848584 3.900 -0.400
39 1 0.85164 40.710 110.258 82.867 -0.225368 -0.023048 0.595161 -0.346745 3.900 -0.400
54 1 0.86112 71.731 96.581 78.999 -0.346745 0.595161 -0.023048 -0.225368 3.900 -0.400
54 1 0.99176 73.711 62.729 97.297 -0.225368 -0.023048 0.595161 -0.346745 3.900 -0.400
54 1 0.91174 51.774 103.448 119.680 0.244501 0.116795 0.417788 -0.779085 3.900 -0.400
56 1 0.96820 45.851 62.446 117.385 -0.346745 0.595161 -0.023048 -0.225368 3.900 -0.400
62 1 0.77175 67.992 66.006 78.669 -0.779085 0.417788 0.116795 0.244501 3.900 -0.400
62 1 0.76489 57.497 120.218 106.229 -0.346745 0.595161 -0.023048 -0.225368 3.900 -0.400
64 1 0.83361 65.631 49.092 92.389 0.244501 0.116795 0.417788 -0.779085 3.900 -0.400
99 1 0.82029 62.630 44.493 70.209 -0.629863 0.543997 0.222109 -0.136243 3.900 -0.400
99 1 0.70661 46.042 94.654 97.889 -0.748573 0.649174 0.649174 -0.549774 3.900 -0.400
101 1 0.80936 83.242 91.310 115.240 -0.629863 0.543997 0.222109 -0.136243 3.900 -0.400
101 1 0.82360 68.789 70.757 94.587 -0.136243 0.222109 0.543997 -0.629863 3.900 -0.400
101 1 0.79289 73.791 122.905 66.823 -0.854901 0.452876 0.050516 0.351509 3.900 -0.400
130 1 0.72926 67.554 60.872 100.415 0.344549 -0.007469 0.305288 -0.642368 3.900 -0.400
130 1 0.74848 52.461 124.421 80.672 -0.833900 0.632008 0.458721 -0.256829 3.900 -0.400
130 1 0.76783 82.299 69.256 56.852 -0.642368 0.305288 -0.007469 0.344549 3.900 -0.400
131 1 0.75497 52.985 75.962 100.165 -0.642368 0.305288 -0.007469 0.344549 3.900 -0.400
145 1 0.89136 82.196 55.199 90.577 -0.833900 0.632008 0.458721 -0.256829 3.900 -0.400
145 1 0.87418 57.042 107.171 117.041 0.351509 0.050516 0.452876 -0.854901 3.900 -0.400
145 1 0.89606 63.280 101.351 75.223 -0.256829 0.458721 0.632008 -0.833900 3.900 -0.400
147 1 0.99519 38.996 72.000 112.916 -0.256829 0.458721 0.632008 -0.833900 3.900 -0.400
153 1 0.73206 65.505 67.541 78.456 -0.854901 0.452876 0.050516 0.351509 3.900 -0.400
155 1 0.84186 64.475 49.574 92.180 0.351509 0.050516 0.452876 -0.854901 3.900 -0.400
155 1 0.69313 53.556 98.903 66.205 -0.629863 0.543997 0.222109 -0.136243 3.900 -0.400
Please refer to the wiki-page of the GENESIS-CG-tool for the meaning of each column.
2. MD simulation of TBP Binding and Induced Bending of DNA
Let’s go to the working directory to carry out MD simulations:
$ cd 02_simulation
$ ls
pro_dna.inp tbp_dna.top tbp_dna_init.gro
2.1 The control file
This is the content of the control file pro_dna.inp:
[INPUT]
grotopfile = tbp_dna.top
grocrdfile = tbp_dna_init.gro
[OUTPUT]
pdbfile = pro_dna_test_run.pdb
dcdfile = pro_dna_test_run.dcd
rstfile = pro_dna_test_run.rst
[ENERGY]
forcefield = RESIDCG
electrostatic = CUTOFF
cg_cutoffdist_ele = 52.0
cg_cutoffdist_DNAbp = 18.0
cg_pairlistdist_ele = 57.0
cg_pairlistdist_PWMcos = 23.0
cg_pairlistdist_DNAbp = 23.0
cg_pairlistdist_exv = 15.0
cg_sol_ionic_strength = 0.15
cg_pro_DNA_ele_scale_Q = -1.0
[DYNAMICS]
integrator = VVER_CG
nsteps = 3000000
timestep = 0.010
rstout_period = 100000
crdout_period = 10000
eneout_period = 10000
nbupdate_period = 100
stoptr_period = 100
[CONSTRAINTS]
rigid_bond = NO
[ENSEMBLE]
ensemble = NVT
tpcontrol = LANGEVIN
temperature = 300
gamma_t = 0.01
[BOUNDARY]
type = NOBC
Most of the options have been already explained in tutorial
11.1 and
11.2. Specifically, here we have one
more “pairlistdist” option for the PWMcos: cg_pairlistdist_PWMcos.
Notably, the charge of phosphate in the 3SPN.2C model is set to -0.6e (for
electrostatics between DNA particles). However, in protein-DNA interactions we
use a value of -1.0e, considering the effect of counter-ion redistribution 3, 6. Therefore, here we set a new value for the phosphate charge in the
electrostatic interactions between protein and DNA to be
cg_pro_DNA_ele_scale_Q = -1.0.
2.2 The topology file
In the same directory we also provided the topology file (tbp_dna.top)
for the protein-DNA complex:
#include "./param/atom_types.itp"
#include "./param/flexible_local_angle.itp"
#include "./param/flexible_local_dihedral.itp"
#include "./param/pair_energy_MJ_96.itp"
; Molecule topologies
#include "./top/dsDNA_cg.itp"
#include "./top/tbp_cg.itp"
[ system ]
TBP-DNA specific interactions
[ molecules ]
dsDNA_cg 1
tbp_cg 1
[ cg_ele_chain_pairs ]
ON 1 - 3 : 1 - 3
OFF 3 - 3
[ pwmcos_chain_pairs ]
ON 1 - 2 : 3 - 3
The first four lines import the general parameters. Then in line 7 and 8 we include the topology files for DNA and TBP, respectively.
Now let’s copy the necessary .itp files to the subdirectory top/:
$ mkdir top/
$ cp ../01_setup/dsDNA_cg.itp top/
$ cp ../01_setup/tbp_cg.itp top/
Let’s go back to the tbp_dna.top. In the [ system ] block we specify the
name for the system. Importatnly, in the [ molecules ] block, we assign the
name and the number of each molecule. Note that the molecule names listed here
should be the same as the ones written in the [ moleculetype ] block of the
“.itp” files. For instance, we can print out the molecule name of the dsDNA in
the dsDNA_cg.itp:
$ head -n 3 top/dsDNA_cg.itp
[ moleculetype ]
;name nrexcl
dsDNA_cg 3
As can be seen, the name of dsDNA is the same as the one we write in the
tbp_dna.top.
Besides, the order of molecules in the [ molecules ] block should be the same
as the order of molecules in the coordinate file (“tbp_dna_init.gro”,
described in the next section).
There are two more blocks describing the inter-molecular interactions, namely,
the “[ cg_ele_chain_pairs ]” and the “[ pwmcos_chain_pairs ]”. In the “[
cg_ele_chain_pairs ]” we first turn on all the intra- and inter-molecular
electrostatics in the system, then turn off the intra-molecular electrostatic
interactions in the TBP (chain No. 3). Similarly, in the “[ pwmcos_chain_pairs
]”, we assign the PWMcos interactions between DNA (chains No. 1 and 2) and the
TBP (chain No. 3). You can find more explanations at the wiki-page of the
GENESIS-CG-tool.
2.3 The coordinate file
The tbp_dna_init.gro file contains all the coordinates of DNA and TBP. The
initial structure was taken from a structure reported in reference 3. In this
structure TBP has already found its consensus sequence on DNA, and the DNA is
bent to an extent similar to the PDB structure. You can start from a dissociated
state, but it may take a long time for the TBP to find its target on DNA.
Therefore, here we simply start from the bound state.
2.4 Run simulation
Now let’s execute atdyn:
$ export OMP_NUM_THREADS=2
$ mpirun -np 4 /home/user/GENESIS/bin/atdyn pro_dna.inp > pro_dna_test_run.log
This simulation will create the following new files:
pro_dna_test_run.dcd: MD trajectory file;pro_dna_test_run.pdb: PDB structure of the last frame;pro_dna_test_run.rst: MD restarting file;pro_dna_test_run.log: GENESIS log file for MD simulation.
You can use VMD or other MD visualization software to take a look at the
structure (pro_dna_test_run.pdb) or the trajectory (pro_dna_test_run.dcd).
The structure of the TBP-DNA complex looks like this:
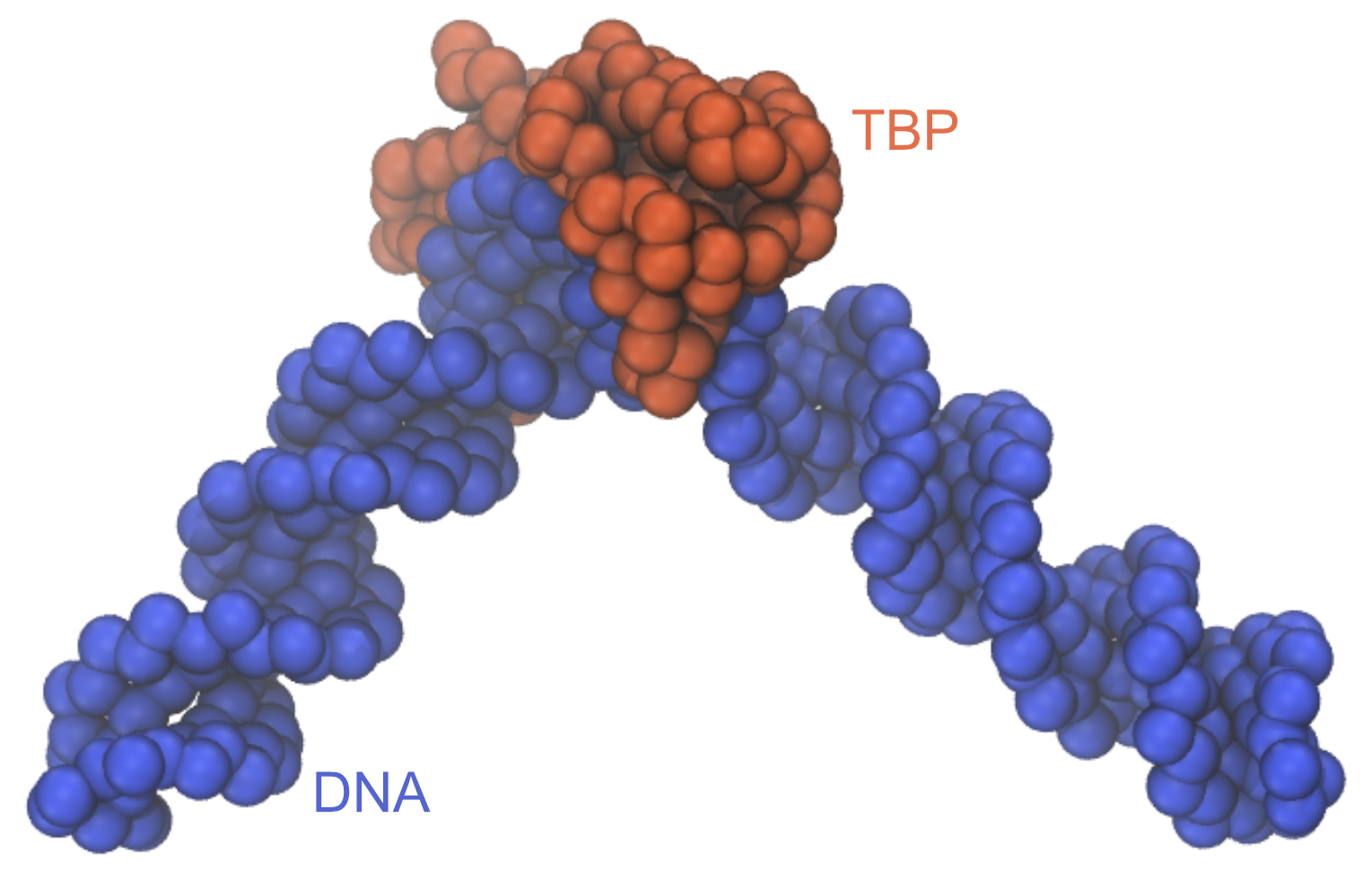
Written by Cheng Tan@RIKEN Center for Computational Science, Computational Biophysics Research Team, October, 2021
References
-
Li W., Wang W., Takada S., 2014, Proc. Nation. Acad. Sci., 111, 10550-10555. ↩
-
Freeman G. S., Hinckley D. M., Lequieu J. P., Whitmer J. K., de Pablo J. J., 2014, J. Chem. Phy., 141 (16), 165103. ↩
-
Tan C., Takada S., 2018, J. Chem. Theor. Comput., 14, 3877-3889. ↩ ↩2 ↩3 ↩4
-
Tan C., Jung J., Kobayashi C., Ugarte La Torre D., Takada S., and Sugita Y., 2022, PLoS Computational Biology 18(4), e1009578. ↩
-
Keishin Nishida, Martin C. Frith, Kenta Nakai, 2009, Nucleic Acids Research, 37 (3), 939–944. ↩
-
Lequieu J., Cordoba A., Schwartz D. C., de Pablo J. J., 2016, ACS Cent. Sci., 2, 660-666. ↩